Antibiotic Resistance
Antibiotic resistance is turning into a global healthcare problem. The exponential growth of metagenomics data has contributed to creation of more accurate and fastest algorithms.
What is DeepARG?
DeepARG is a machine learning solution that uses deep learning to characterize and annotate antibiotic resistance genes in metagenomes. It is composed of two models for two types of input: short sequence reads and gene-like sequences.
Short NGS reads
DeepARG is able to annotate short sequence reads from Next Generation Sequencing (NGS) technologies such as Ilummina. This model has been trained with simulated antibiotic resistance reads to perform better with metagenomic samples.
Gene-Like Sequences
DeepARG is able to predict antibiotic resistance in long gene-like sequences. This model is suitable for annotating full sequence genes and to discover novel antibiotic resistance genes from assembled samples.
Documentation
Antibiotic resistance Database
Please visit our repository to download deepARG-DB
Important! DeepARG-DB has been under continuous inspection in order to improve the quality of the models. As result, we created a new online resource called ARGminer that allows the manual inspection of ARGs.
For more information please visit the ARGminer website
Train Your deep Learning Model
We have included the neccesary steps to re-train the deepARGs models or to create your own deeep learning model using the architecture of deepARG.Please look at the Bitbucket README file for details in the DeepARG repository.
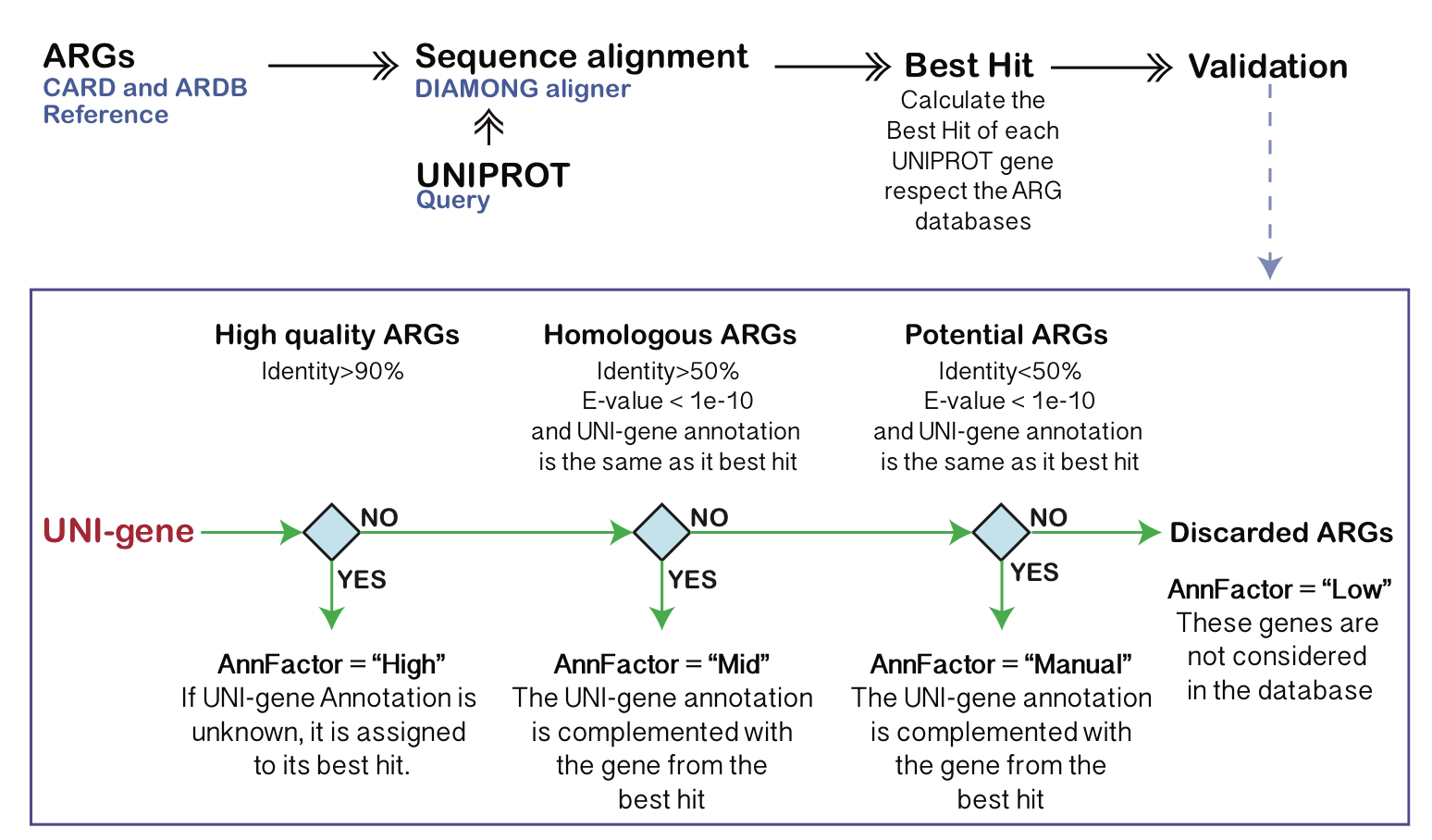
Figure 1: Automatic annotation of highly homologous ARGs
Deep Learning Models
The deepARG models have been designed for computational analysis of next generation sequencing data such as Metagenomes. The main contribution of the deepARG models are their low false negative rate during predictions. Also, the gene-like sequences model is designed to find novel ARGs based on the sequence homology.
DeepARG pipeline
The pipeline can be used as an stand alone program. It was developed in python 2.7 and requires (optional) DIAMOND for making the alignments. The source code can be downloaded from this Git Repository hosted in BitBucket.
Dependencies
DeepARG requires the next python modules (all can be installed via pip):
- Nolearn lasagne deep learning library.
- Sklearn machine learning routines.
- Theano for fast computation. For GPU usage (see theano documentation)
Instalation
Open a terminal and clone the source code:
git clone https://bitbucket.org/gusphdproj/deeparg-ss
Configuration
-
Go to the directory where the program was saved and open the file
options.pyReplace
path = '/home/gustavo1/tmp/deeparg-ss/';with the current directory (deepARG path).
For instance, deepARG was cloned at/home/user/deeparg-ss/
The options.py file should looks like
path = '/home/user/deeparg-ss/'; -
Go to
./binunder deeparg-ss and runchmod +x diamond(only for LINUX)
Usage
Please visit our repository for details
Direct Annotation Service
New! (9/6/2017) We are excited to release a new version of our deepARG online analysis.
Meet our projects on metagenomics analysis
Visit Website
MetaStorm incorporates several functional databases such as ACLAME, COG, UNIPROT, CARD, ARDB, BACMET among others.
Visit Website
Visit Website
How to cite Us
Funding
HEARD: NSF Halting Environmental Antimicrobial Resistance Dissemination.
The Virginia Tech Sustainable Nanotechnology Interdisciplinary Graduate Education Program (IGEP).
Our team Who is behind this project
Contact us Keep in touch
The easiest and preferred way to get in touch with our team is by posting an issue, question or comment to our ARGminer blog
For instance if you are interested on knowing how to run deepARG using the docker contanier version, you can find a tutorial here: DeepARG docker execution
Or you can post any question regarding the deepARG usage as input for deepARG local version
We strongly recommend to contact us via our blog as other users may find your questions, issues or comments useful too.
You can still send us an email using the form below
- Address
- Virginia Tech, VA 24061
- Phone number
- (+1) 202 717 5300
- lqzhang@cs.vt.edu
 Department of Computer Science
Department of Computer Science
 Department of Civil and Environmental Enginneering
Department of Civil and Environmental Enginneering
 Advanced Computing Center (ARC)
Advanced Computing Center (ARC)
